Less than a year ago (March 11 to be precise), the Director General of the World Health Organization (WHO), Dr. Tedros Adhanom, declared that COVID-19 could begin to be classified as a pandemic.
Given this, the relative search volume (RSV) of the new coronavirus soared and the consumption of information regarding it became massive. Sadly, in the same vein Huxley warned us, we gorge ourselves on irrelevance and misinformation. Pseudo-knowledge began to arrive without major filters and the propagation of misinterpreted data, nonsense and fantastic inferences became entrenched in the popular imagination under the protection of figures who projected security at the given time.
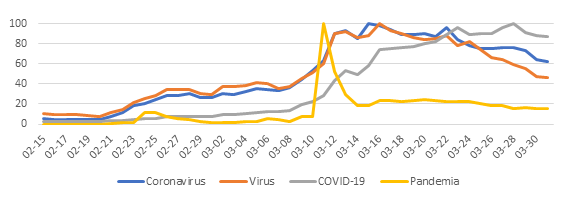
At the community level, it was foreseeable that the uncertainty and inconsistency of data would generate fear, frustration and damage our perception of self-efficacy to face the situation we were entering. Therefore, it is not surprising that the health sector and academia are now facing a series of myths and inaccuracies regarding SARS-CoV-2 and the protective actions to avoid its contagion.
COVID-19 in the field of veterinary medicine
A temporal trend analysis for the search of health information indicates that there is some concern in the population about the capacity of domestic animals to contract and spread the disease (Figure 2).
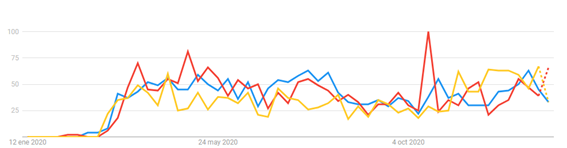
For this reason, and so because we want you to base your recommendations on these and other inquiries on science and evidence, we have prepared a series of articles where you can find everything you need to know about SARS-CoV-2, COVID-19 and their implications for the veterinary medicine industry, as well as a review of phylogenetically close diseases that affect animals.
Without further ado and inspired by “Sun Tzu’s Art of War” as an information management scheme, we began the process to better understand this disease and the virus that causes it.
Differences between COVID-19 and SARS-CoV-2
Although you probably already know, it is important to be clear:
- COVID-19: the disease caused by the virus (Coronavirus Viral Disease – 2019)
- SARS-CoV-2: the etiological agent of the disease.
In other words: The presence of this virus in a person or animal is called SARS-CoV-2 infection, while the development of disease caused by the virus (regardless of symptoms or severity) is called COVID-19.
Why is this clarification important?
The presence of a virus in any organism does not always have clinical implications. The development of a disease depends on the degree of affection that the virus can generate on the physiological state of the host.
Sometimes the replication of a virus in a host does not result in serious or relevant histological lesions, in the case of SARS-CoV-2, the reasons for an infection without symptoms are still not fully understood.
The matter is still in the field of speculation but it is proposed that the severity of the symptoms could be dose dependent and that the adaptation of the virus to ACE2h receptors is not yet complete (the new strains of which we are having knowledge mutated precisely in the receptor-mandatory domains).
Origins of SARS-CoV-2
SARS-CoV-2, initially called 2019-nCoV, was discovered by Dr. Zhu’s research group in samples from patients hospitalized in Wuhan between December 2019 and January 2020.
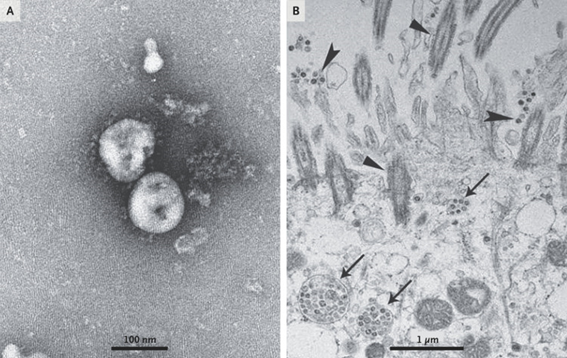
Although this is ignored by some media and communicators, the virus itself was isolated before the pandemic started. In fact, 3 cell lines were used for this purpose: human airway epithelial cells, Vero E6 and Huh-7.
All the cell cultures in that first study showed cytopathic effects with the difference that the cells of the respiratory epithelium only took 92 hours while the rest 6 days after inoculation. These cultures were the ones that made possible the collection of virions for their structural characterization by transmission electron microscopy.
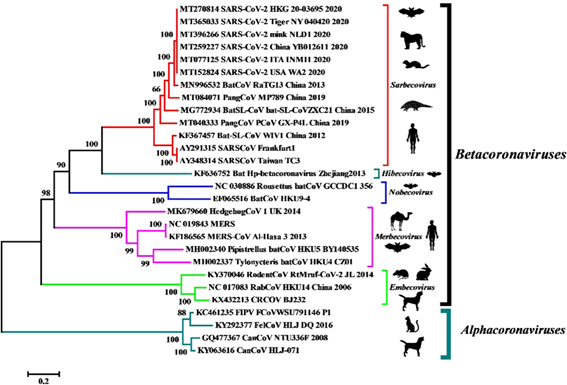
The molecular identification of the virus was given by means of the metagenomic study of the RNA of samples of the alveolar bronchial lavage of the patients and analyzed the more than 10 million reads obtained from the illuminia and nanopore platforms from which it was later possible to reconstruct a complete genome of 29891 base pairs with 79.6% identity to SARS-CoV BJ01. Subsequent bioinformatic analysis revealed that the closest common ancestor that we would be aware of is RaTG13, a beta-coronavirus discovered in bat feces in 2013.
Although it only retains 1 of the 6 important residues for the receptor-binding domain of the S gene, RaTG13 also uses angiotensin-converting enzyme 2 (ACE-2) as a cellular input and maintains 90.55% identity with the second relative. closest, the pangolin coronavirus (Pangolin-CoV MP789).
This other coronavirus, despite having only 91.02% identity at the whole genome level with SARS-CoV-2, exhibits perfect consistency with the 6 residues that maintain physical contact during interaction with human ACE-2 receptors, giving us the idea that could be involved in the evolutionary origin of the pandemic.
Regarding the anthropogenic origin of SARS-CoV-2 as a laboratory accident or under genetic manipulation, we believe that, although it is unlikely, the discussion should continue to clarify any doubts in this regard.
Moving away from speculation, for now we know that:
- BatCoV RaTG13 (MN996532) and PangCoV MP789 (MT084071) are the closest relatives of SARS-CoV-2.
- RaTG13 does not bind to pangolin ACE-2.
- Chimeric CoVs have been created before.
- Until 2020 the study of CoVs only required a level 2 biosafety laboratory.
- SARS-CoV-2 differs by more than 6000 nucleotides from SL-SHC014-MA15 (a chimeric CoV that acquired increased pathogenicity in mice and its study was restricted under the policy of gain of function).
- The portions of the genome that SARS-CoV-2 shares with HIV-1 are not specific and the initial report from the New Delhi Institute of Technology has already been retracted.
Susceptible species
Predictive modeling based on ACE-2 sequences from other mammals and previous studies with SARS-CoV-1 made it possible to temporarily propose species susceptible to infection and these were later confirmed experimentally to detect reservoirs and models. animals that can help us with your research.
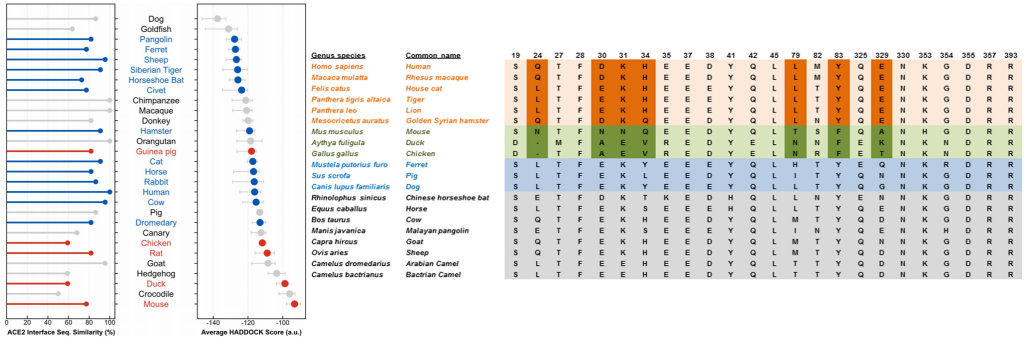
To date, officially susceptible animals that may develop symptoms are:
- Non-human primates (Macaca mulatta, Macaca fascicularis, Chlorocebus sabaeus)
- Malacian Tigers (Panthera tigris jacksoni)
- Siberian Tigers (Panthera tigris altaica)
- African lions (Panthera leo)
- American mink (Neovison vison)
- Ferrets (Mustela putorius furo)
- Golden Hamsters (Mesocricetus auratus)
- Dogs (Canis lupus familiaris)
- Cats (Felis silvestris catus)
From this list, only in American minks has it been possible to identify a variant capable of making a jump from host to human, which is why the Danish government decided to slaughter more than 17 million minks in 2000 farms and terminate its participation in the fur industry, an activity for which it had a turnover of around half a billion euros per year.
The probability of cats and dogs developing the disease is low and, according to the reports deposited in the OIE’s World Health Information System (WAHIS), the morbidity would not be severe and their mortality would be associated more than anything with other debilitating diseases.
In the case of dogs, there does not seem to be the possibility of contagion between specimens of the same species or other animals, a different scenario from cats and ferrets in which it is possible that they could infect other animals of the same species, although it has been already hypothesized its involvement in the spread of the virus in Danish mink farms.
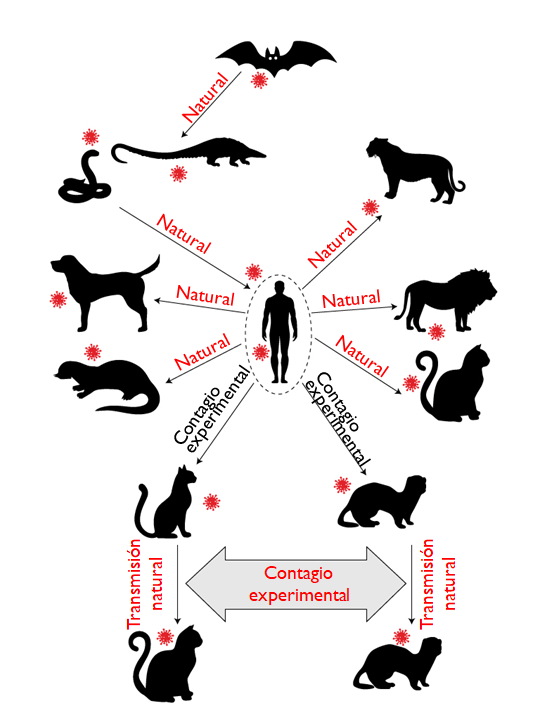
Figure 6. Ability of SARS-CoV-2 to infect domestic animals. Adapted from Hossain et al., 2020
Later on, we’ll be talking about other CoVs of veterinary interest, their pathogenesis and the state of the art in their treatment.
Send us your opinion through the comments or write to the following email.
If you liked this article, subscribe and share it on your social media.
Bibliography
- Borkovich D., Noah P. 2014. Big Data in the Information Age: Exploring the Intellectual Foundation of Communication Theory. Information Systems Education Journal (ISEDJ) 12 (1), 15-26.
- Borkovich, D. J. (2012). When corporations collide: Information overload. Issues in Information Systems, 13(2), 269-284.
- https://www.nejm.org/doi/pdf/10.1056/NEJMoa2001017?articleTools=true
- Phan, T. (2020). Novel coronavirus: From discovery to clinical diagnostics. Infection, Genetics and Evolution, 104211.
- Zhou P, Yang X-L, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. (2020) 579:270–3. doi: 10.1038/s41586-020-2012-7
- Tang X, Wu C, Li X, Song Y, Yao X, Wu X, et al. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. (2020) 7:1012–23. doi: 10.1093/nsr/nwaa036
- Zhang, T., Wu, Q., & Zhang, Z. (2020). Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Current Biology.
- https://www.cdc.gov/sars/guidance/f-lab/app5.html
- Rodrigues, J. P. G. L. M., Barrera-Vilarmau, S., M C Teixeira, J., Sorokina, M., Seckel, E., Kastritis, P. L., & Levitt, M. (2020). Insights on cross-species transmission of SARS-CoV-2 from structural modeling. PLoS Computational Biology, 16(12), e1008449.
- Kuo, L., Godeke, G. J., Raamsman, M. J. B., Masters, P. S., & Rottier, P. J. M. (2000). Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: Crossing the host cell species barrier. J. Virol., 74, 1393‐1406.
- Maier, H. J., Bickerton, E., & Britton, P. (2015). Coronaviruses – Methods and protocols. London: Humana Press.
- Becker, M. M., Graham, R. L., Donaldson, E. F., Rockx, B., Sims, A. C., Sheahan, T., … Denison, M. R. (2008). Synthetic recombinant bat SARS‐like coronavirus is infectious in cultured cells and in mice. PNAS, 105, 19944‐19949.
- Hu, B., Zeng, L. P., Yang, X. Lou, Ge, X. Y., Zhang, W., Li, B., … Shi, Z. L. (2017). Discovery of a rich gene pool of bat SARS‐related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog., 13, 1‐27.
- CsiszarA, JakabF, ValencakTG, LanszkiZ, TóthGE, KemenesiG, TarantiniS, Fazekas-PongorV, UngvariZ. 2020. Companion animals likely do not spread COVID-19 but may get infected themselves. Geroscience. 42(5):1229–1236.
- EnserinkM. 2020. Coronavirus rips through Dutch mink farms, triggering culls. Science. 368(6496):1169.